Precambrian Protein
August 26, 2013
Money is a problem in most organizations, and that includes
research organizations. As a consequence,
scientists use twenty-year-old
voltmeters and
oscilloscopes because they're the only ones they have. One of my
technicians would often recite the following
adage while working in the
lab: "
If it ain't broke, don't fix it." You should never pry the cover from old, working
equipment. If you do, it might not work when you put the cover back on.
Nature seems to believe the same when it comes to
evolution. When evolution created a working design for a
limb, it was applied to
fins,
wings and
legs. According to today's evolutionary
framework, the
tetrapod limb evolved from the fish fin, and bird wings evolved from the tetrapod limb. It's not surprising to see
pentadactyly (from the Greek, πεντε, five, plus δακτυλος, finger) preserved among different
species.
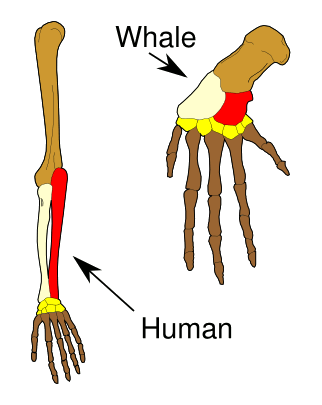
Darwin argued that the similarity of body parts between species (homology) was indicative of a common ancestor, since it's apparent that the particular skeletal structure of the limb, for example, is not the best possible structure for its different function in different animals.
(Illustration by Volkov Vladislav Petrovich, via Wikimedia Commons.)
Evolution seems to have taken the same advantage of the tried and true at the
chemical level, as recent research on the
protein,
thioredoxin, indicates. An international research team from the
Universidad de Granada,
Columbia University and the
Georgia Institute of Technology provides evidence that only small
structural changes have occurred in this protein over four billion years from the time of their
last common ancestor.[1-3]
Thioredoxin is an
enzyme having several
metabolic functions in
cells, since it can break
sulfur bonds in
molecules. This enzyme is used by nearly every species, from
bacteria to
humans; and this indicates that the single-celled ancestor of life would have had
genes for production of this protein.[2]
To back-track the evolution of these genes, thioredoxins were sampled from a wide variety of modern
organisms, analyzed, and placed in an evolutionary context. The resulting
phylogenetic tree was derived by analyzing the
amino acid sequences of thioredoxins in the various species (see figure). This phylogenetic tree gave the information needed to resurrect the most ancient protein, dating from the
Precambrian era, and characterize its properties.[3]
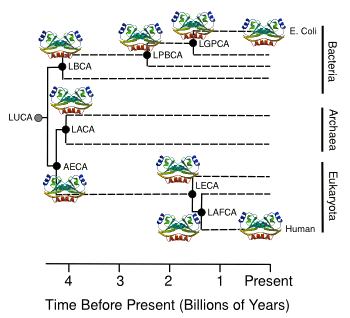
The phylogenetic tree for the evolution of thioredoxins.
(Illustration by the author using Inkscape from data in ref. 1.[1] Protein structure illustration via Wikimedia Commons.)
Senior author,
Jose Sanchez-Ruiz of the University of Granada, said that the approach used "...comes as close as possible to 'digging up' fossil protein structures."[3] Computer analysis was used to predict the
DNA sequences going back as far as four billion years ago.[2] The analysis was aided by the fact that
protein biosynthesis would not happen when as few as one or two genes are out of place. Explains coauthor,
Eric Gaucher, a
professor at Georgia Tech,
"If our ancestral sequences were incorrectly inferred by having a single mistake, that could have led to a dead gene. Instead, our approach created biochemically active proteins that fold up into three dimensional structures that look like modern protein structures, thus validating our approach."[2]
The research team then inserted the protein synthesizing genes into bacteria to produce quantities for analysis.[2]
Earth's environment four billion years ago was extreme, with high
temperatures and highly
acidic water.[2] The study found that the ancient protein was quite hardy, surviving at temperatures of more than 110°
C. It was also stable in
acid.[2] The ancient ancestor of life on Earth was an "
extremophile," a bacteria like those of today living in places such as
hot springs.[2]
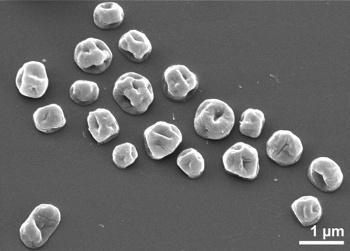
S. hellenicus, a representative extremophile.
This bacteria grows between 70-90°C in a solution pH of 4.5-7.5.
(Image via Open-i, the Open Access Biomedical Image Search Engine.)
Although the amino acid sequences are very different, present day thioredoxins are very similar to that of the Precambrian thioredoxin.[3] The authors remark that this finding supports a
punctuated-equilibrium model of evolution. In that model, protein structure remains constant over long periods, and changes occur intermittently over short periods.[2-3]
References:
- Alvaro Ingles-Prieto, Beatriz Ibarra-Molero, Asuncion Delgado-Delgado, Raul Perez-Jimenez, Julio M. Fernandez, Eric A. Gaucher, Jose M. Sanchez-Ruiz and Jose A. Gavira, "Conservation of Protein Structure over Four Billion Years," Structure, August 8, 2013, DOI:10.1016/j.str.2013.06.020.
- Simon Redfern, "Resurrected protein's clue to origins of life," BBC News, August 8, 2013.
- 'Digging up' 4-billion-year-old fossil protein structures to reveal how they evolved, Press Release, Cell Press, August 8, 2013.
Permanent Link to this article
Linked Keywords: Research; scientist; voltmeter; oscilloscope; technician; adage; laboratory; lab; If it ain't broke, don't fix it; scientific instrument; equipment; Nature; evolution; limb; fish fin; wing; leg; working hypothesis; framework; tetrapod; dactyly; pentadactyly; species; Charles Darwin; homology; common descent; common ancestor; skeleton; skeletal; Wikimedia Commons; chemical compound; protein; thioredoxin; University of Granada; Universidad de Granada; Columbia University; Georgia Institute of Technology; protein structure; last common ancestor; enzyme; metabolism; metabolic; cell; sulfur; chemical bond; molecule; bacteria; human; gene; organism; phylogenetic tree; peptide sequence; amino acid sequence; Precambrian era; Inkscape; Jose Sanchez-Ruiz; nucleic acid sequence; DNA sequence; protein biosynthesis; Eric Gaucher; professor; Earth; environment; temperature; acidic; water; Celsius; C; acid; extremophile; hot springs; Staphylothermus; S. hellenicus; pH.