The Human Microbiome
June 18, 2012
The
microbiome is the ensemble of
microbial communities that live on, and in, the
human body. Obviously, something that's living and in such intimate contact with us should have some impact on our
health. the
National Institutes of Health launched a
Human Microbiome Project (HMP) in 2008 to study the microbiomes of 242 healthy
humans.
(NIH Image)
is not always that the microbes cause the disease. An analysis of the microbiome may be a predictor of disease. To such an end, the HMP is developing a reference set of
genome sequences for the microbiome, and establishing a repository. The project goal is to sequence 3000
genomes to at least a high-quality draft stage. As of this writing, about 25% of this goal has been achieved.
The ~200 members of the Human Microbiome Project from nearly 80
research institutions have just published a slew of
papers describing their results.[1-9] There were fourteen papers published, two of which were in the June 14 issue of
Nature[1-2] and the remaining twelve in
Public Library of Science (PLoS) journals.[3-4]
These papers describe the analysis of more than 5,000 samples of human and bacterial
DNA that resulted in 3.5
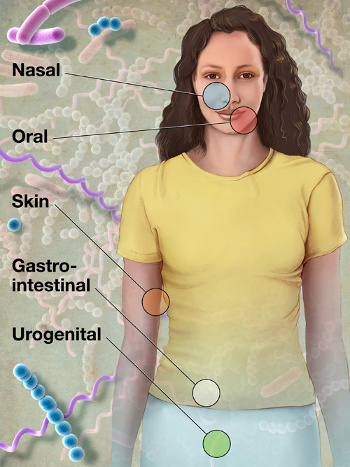
terabases of genomic data.[5] Samples were obtained from the the airways, skin, oral cavities, digestive tracts, and vaginas of 242 healthy people ranging from 18 to 40 years old and living around
Houston or
St. Louis.[4]
It's estimated that the number of microbes in our microbiome outnumber our
human cells by a factor of ten.[4] The HMP concludes that there are about 10,000 bacterial species in our human microbiome.[5,7] These microbes are thought to contain eight million different
protein-coding genes. The human genome contains about 22,000 protein-coding genes, so the microbial protein factories are 360 more diverse.[4]
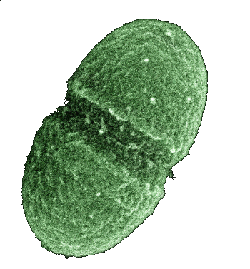
An Enterococcus faecalis bacterium, normally found in the human gut.
(United States Department of Agriculture image))
One interesting fact is that the microbial distribution in the human body is similar to that of the
ocean ecosystem,[5] but the difference in the microbiome from person to person can be extreme. The common bacterium,
Bacteroides, might comprise nearly 100% of the microbes in one person's gastrointestinal tract, but it may be completely absent in another person.[5]
Susan M. Huse of the
Marine Biological Laboratory summarizes this finding by saying,
"What this means is, there is not just one way to be healthy... There doesn’t have to be one or two 'just right' gut communities, but rather a range of 'just fine' communities."[5]
Racially and ethnically related people seemed to have similar microbiomes. However, the differences in microbiome composition didn't matter at a functional level, since the set of microbes in each person performed similar
metabolic tasks.[4]
The HMP researchers found also that most people carry
pathogenic microbes without ill affect. This leads to the problem as to what conditions will lead to disease.[5]
Staphylococcus aureus, one strain of which is the difficult to treat
Methicillin-resistant Staphylococcus aureus, was found in the noses of about 30 percent of the people.[4]
Dirk Gevers, who shared first authorship of one Nature paper,[1] and was an author of the other,[2] compared the Human Microbiome Project to the
Human Genome Project.
"Just as the Human Genome Project was 10 years ago, the Human Microbiome Project is intended to be a baseline for future studies of human health and disease... This is a tremendous resource that is now publicly available to the scientific community that allows us to ask how and why microbial communities vary."[4]
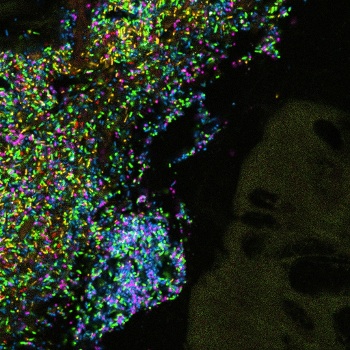
An Intestinal section from a gnotobiotic mouse model inoculated with selected bacterial species found in the human gut. Blue=Bacteroides WH2, green=Bacteroides thetaiotamicron, pink=Bacteroides vulgatus, yellow=Collinsella aerofaciens, red=Ruminococcus torques.
(Image: Yuko Hasegawa/MBL Woods Hole))
The Human Microbiome Project is managed by the
National Human Genome Research Institute (
http://www.genome.gov).
References:
- Curtis Huttenhower, et al. (The Human Microbiome Project Consortium), "Structure, function and diversity of the healthy human microbiome," Nature, vol. 486, no. 7402 (June 14, 2012), pp. 207-214; PDF file available, here.
- Barbara A. Methé, et al. (The Human Microbiome Project Consortium), "A framework for human microbiome research," Nature, vol. 486, no. 7402 (June 14, 2012), pp. 215-221; PDF file available, here.
- Susan M. Huse, Yuzhen Ye, Yanjiao Zhou and Anthony A. Fodor, "A Core Human Microbiome as Viewed Through 16S rRNA Sequence Clusters." PLoS ONE, vol. 7, no. 6. Document No. e34242; PDF file available, here.
- Elizabeth Cooney, "Mapping the healthy human microbiome," Broad Institute Press Release, June 13, 2012.
- 'More than One Way to Be Healthy': First Map of the Bacterial Makeup of Humans Relies on MBL Innovations, Insights, Marine Biological Laboratory Press Release, June 13, 2012.
- NIH Human Microbiome Project defines normal bacterial makeup of the body, National Human Genome Research Institute (NHGRI) Press Release, genome.gov June 13, 2012; PDF version available, here.
- Anne Holden, "Consortium of Scientists Map the Human Body's Bacterial Ecosystem," Gladstone Institute Press Release, June 13, 2012.
- Forsyth Scientists Define the Bacteria that Live in the Mouth, Throat and Gut, Forsyth Institute Press Release, June 13, 2012.
- Human Microbiome Project, The NIH Common Fund web site.
Permanent Link to this article
Linked Keywords: Microbiome; microorganism; microbial; human body; health; National Institutes of Health; Human Microbiome Project; humans; bacteria; fungi; archaea; skin; mouth; eye; gastrointestinal tract; woman; vagina; intestine; intestinal; digestion; carbohydrate; NIH; disease; genome sequence; genome; research institution; scientific literature; Nature; Public Library of Science; DNA; terabase; Houston; St. Louis; human cell; protein; Enterococcus faecalis bacterium; United States Department of Agriculture; ocean; ecosystem; Bacteroides; Susan M. Huse; Marine Biological Laboratory; metabolism; metabolic; pathogenic bacteria; pathogenic microbe; Staphylococcus aureus; Methicillin-resistant Staphylococcus aureus; MRSA; Dirk Gevers; Human Genome Project; gnotobiotic; mouse; Collinsella; Ruminococcus; Yuko Hasegawa; MBL; Woods Hole; National Human Genome Research Institute; www.genome.gov.